3D Segmentation by Deformable M-reps
- Recent IPMI'2001 publications related to M-Reps modeling:
- Intuitive, Localized Analysis of Shape Variability
- Multi-scale 3-D Deformable Model Segmentation Based on Medial Description
- Medial models incorporating object variability for 3D shape analysis
- Object-based image analysis at multiple scales: TBD
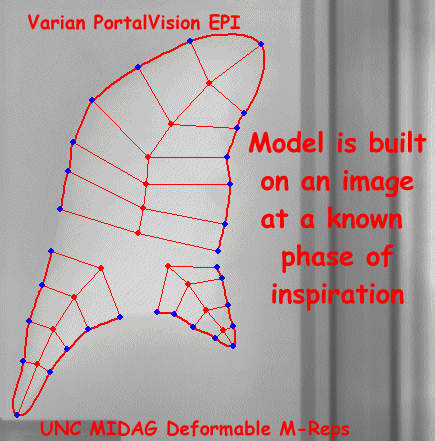
Radiotherapy Treatment Planning via Deformable M-reps Segmentation
- 2D Automatic Segmentation of Lung in Electronic Portal Image
- Segmentation of Single-Figure Objects by Deformable M-reps
 |
Note: you may click on this image to play the movie but GIF animations play faster and smoother if you copy it to your local disk and play it from there. Use "Save Target As..." or "Save Link As..." |
Statistical Shape Characterization for Neuroscience
- Shape of subcortical brain structures in schizophrenia from MRI: New results and background data
- Recent IPMI'2001 submissions related to M-Reps modeling:
- Medial models incorporating object variability for 3D shape analysis
|
Statistical analysis of lateral ventricles in Mono/Di-zygotic twins, volume and shape |
 |
|
|
|
|
|
 |
|
|
|
| Gerig, G, M Styner and J Lieberman (2001). Shape versus Size: Improved understanding of the morphology of brain structures. Proc. MICCAI2001, to appear October 2001, Kluver. |
(extended Technical report version)